What is a grape variety? What is a clone? Unpacking the science
We talk a lot about clones and grape varieties. But what, precisely, do these terms mean?
Let’s first define ‘variety’. A new grape variety is produced when a grape seed has grown into a plant. So the 1500 or so commercial varieties of the single species Vitis vinifera grown worldwide are all the result of the germination of 1500 different seeds. Examples? Pinot Noir, Cabernet Sauvignon, Chardonnay, Sauvignon Blanc, Merlot, Syrah. Each grape variety we have now consist of plants that are descended from that single seed, and which have been selected by humans, and have then been propagated over centuries or millennia by people layering the vines or taking cuttings (known as vegetative propagation, as opposed to sexual propagation).
As Jose Vouillamoz, the grape geneticist puts it: every variety has a father and mother. That’s all you need to make a new variety, and it’s also the only way you can make a new variety. If you take a seed from a Chardonnay grape, and grow it, you will have a new variety. But if, over hundreds of years, people take cuttings from Chardonnay grapes and keep on propagating them, they will remain Chardonnay.

Over time, as grape vines of each variety are propagated vegetatively, spontaneous mutations will accumulate in their DNA. One of the sources of this mutation is the copying process itself: each time a cell divides by mitosis (as opposed to meiosis which occurs during the production of pollen or ovules in sex), a new strand of DNA is copied from the existing one. There are molecular checks and balances, but occasionally mistakes are made. Then there are environmental stresses that can cause DNA mutations. These vines are standing out in a field, often in very sunny climates, and will be exposed to a lot of solar radiation. The sorts of changes that can occur include single base changes in the DNA code, or insertions or deletions of bits of DNA (called ‘indels’: these are structural changes) or genes moving around (known as transposable elements; TEs). TEs have been shown to be responsible for most clonal variation in grape varieties, and form around 20% of the DNA sequences in Vitis vinifera. Their activity is heightened by environmental stress: it is becoming clear they are important for plant adaptation to the environment. Finally, there’s epigenetic modification, which involves changes to the material around DNA but not the DNA sequence itself, affecting how genes are turned on and off, and this can be heritable just as DNA changes are heritable. Epigenetic changes also allow a vine to adapt to the environment, partly in concert with the TEs.
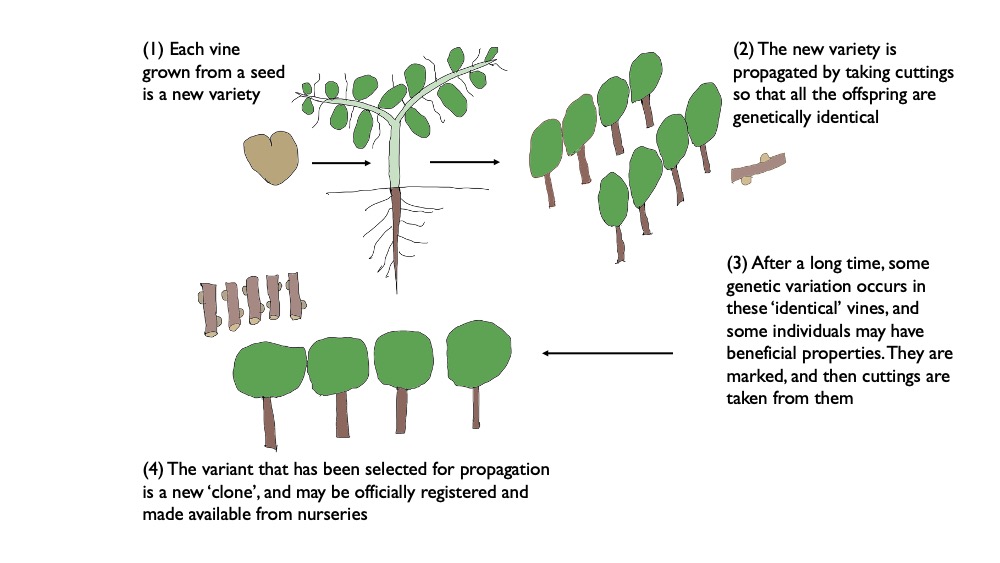
If you have 10 000 vines growing in a vineyard of the same variety, each starting out genetically identical, over the years various mutations will develop and there will be some genetic differences among the vines. Most of the mutations will be invisible, and most will be neutral or deleterious. If they occur in a cell that isn’t part of a bud meristem (the area of rapid cell division that forms new bits of plants) they are incidental because they won’t be passed on through cuttings. But if they occur in the bud meristem they can produce a shoot that has different properties to the rest of the vine. So very occasionally these mutations will be visible, and some of these visible changes will be desirable. When it comes to taking cuttings for propagating, some vines might be just a little better than their neighbours, and if this is because of subtle genetic differences (rather than, for example, small differences in terroir that then influence the vine’s growth), then these will be captured by the propagation process.
This sort of selection results in a new clone of the variety. Over enough time, a range of such clones might be developed. But the accumulation of mutations can’t produce a new grape variety. You might have several clones of Pinot Noir developing each with quite different characteristics, but they will remain Pinot Noir. A new variety is only produced with a mother and a father (vine sex), resulting in a seed that is then grown. As an aside, people often say that Pinot Noir is genetically prone to mutation because there are so many different clones. But there is no evidence for this. The variation within a variety increases with the length of time the variety has been grown (Pinot Noir is an ancient variety), the area under vine (this is a numbers game and Pinot Noir has been widely grown) and also whether or not people have actually been looking for the variation and capturing it in clones.
There’s an interesting point here. You could take the parents of a particular variety, where they are known, and cross them, but you wouldn’t get the same variety from this cross. Let’s use Chardonnay as an example. Its parents are Pinot and Gouais Blanc. If you cross Pinot and Gouais Blanc, however, you won’t get Chardonnay. That happened once, but won’t happen again (other varieties including Aligoté, Melon de Bourgogne and Gamay share these parents, and they are all quite different). You’ll get a new variety: the genetic mixing up that took place to produce Chardonnay was a unique event; in a similar way, I’m different to my sisters and brother, and even if my parents had had 3000 children they would never have had me again. [As an aside, if you cross Pinot Noir with Pinot Noir and grow a seed from this cross, you won’t still have Pinot Noir. There will be some genetic mixing up in the sex process that will result in a new variety.]
By this definition of variety and clone, Pinot Noir, Pinot Gris, and Pinot Blanc are not separate varieties. They are genetically indistinguishable by all DNA marker-based tests that are used to distinguish different grape varieties. “Pinot is one single grape variety that is very old and that has had many ‘accidents’ in its life—many mutations,” says Jose Vouillamoz. “Some of them were spectacular because they touched the color of the grape varieties and almost nothing else. When I do the DNA profiling of Pinot Noir, Pinot Gris, and Pinot Blanc, since I look at say 10 or 12 different DNA regions, I don’t see where the color mutation has happened. They are all the same in the DNA profile.”
How can clones be distinguished then? The only way is by sequencing the entire genome. One project aiming to do this is currently taking place in South Africa and the Loire focusing on Chenin Blanc. Johan Burger leads the South African side of things, with the French research group led by Patrice This. They are using next generation sequencing to try to tease apart clones, by looking at the entire genome without the use of predefined markers. Similar work has been done with Chardonnay in Australia, where 1620 markers can distinguish 15 Chardonnay clones and have confirmed Gouais Blanc and Pinot Noir as the parents. With Zinfandel in California a research group has assembled the genome and are sequencing 15 clones. In Spain they are working on Malbec in a similar way. The aims of this Chenin project are to study the accumulated mutations in the different clones, and then develop a sort of genetic assay.
Patrice This, the French partner in the project, started working on grapevine genetic diversity almost 30 years ago, and he’s keen to be involved in a project comparing the evolution of clonal diversity in both France and South Africa. ‘Clonal variation only occurs after extreme replication of material,’ he says, ‘when you can see morphological differences due to sequence variation or epigenetic modifications.’

The grapevine reference genome is PN40024 which was sequenced in 2007. To make the job of sequencing easier, the researchers in Colmar (Alsace) backcrossed Pinot Noir with itself lots of times, so this PN40024 can’t be called Pinot Noir (remember: each time a vine has sex like this, even if it is with the same variety, a new variety is created). Fortunately, sequencing techniques have moved on a lot since then, and now it is much cheaper and easier to sequence the entire DNA of the grapevine. The way this is being done in this Chenin project is to extract DNA from the leaves (this is easier), and then cut it into long fragments using Sequel II (mean size 20 000 base pairs). These fragments are then sequenced, and are aligned to their correct spots using the reference genome. This is still quite a computational task, because the grapevine genome consists of 500 million base pairs spread over 19 chromosomes. The chromosomes come in pairs, and so there are two haplotypes that have to be aligned. This is because genes come from both parents, Savagnin and Sauvignonasse. This creates a reference genome to compare clones with. The next step is to sequence the clones for comparison, and this time the DNA is extracted and then sequenced in short fragments using Illumina. The mean size of these is 150 base pairs, so there are a lot of them. These are then aligned on the reference that has just been made, and the clones are compared at different positions. The work is ongoing.
The fact that the Pinot group aren’t technically separate varieties raises an interesting question. Scientifically, we have a watertight definition of a variety: a vine grown from a seed. But functionally, we can’t live with this in the wine world when it comes to Pinot. Skin colour mutations are almost always clonal differences caused by transposable elements or genetic deletions, but most people can’t get their heads around this and automatically consider them new varieties. And, in practice, it would make no sense from a wine point of view to consider Pinot Noir and Pinot Gris to be the same variety. Legally, the Pinot members aren’t interchangeable, either. Try putting Pinot Blanc into AOC red Burgundy, and you’ll have a tough time arguing your case. So in this case we may have to make an exception. Another example of this is Pinot Meunier, now being called Meunier in Champagne. Once again, this is a new clone of Pinot Noir; it’s just that it’s a chimera, with the epidermal layer of cells (the L1 layer; everything else is from the L2 layer) containing a mutation that affects the production of a plant hormone called gibberellin. This is responsible for the hairy appearance of Pinot Meunier. Normal DNA marker techniques can’t tell Pinot Noir and Pinot Meunier apart.
Overall, though, this scientific definition of clone and variety makes things a lot clearer.
[The picture at the top is of Mendoza clone Chardonnay, which is closely related to another clone called Gingin. Both of these clones are famous for their tendency to show hen and chicken bunch characteristics (millerandage), with small and larger berries, resulting in intensely flavoured wines. This picture was taken at Ata Rangi in New Zealand’s Martinborough region.]